
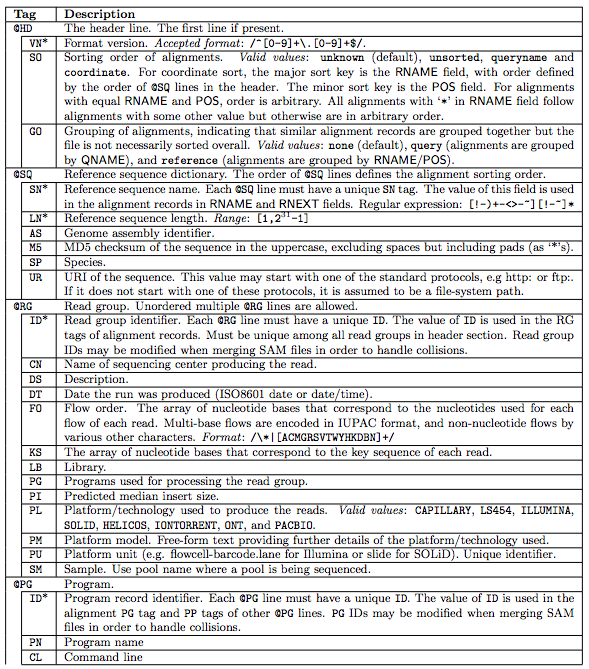
samtools view -C -T ref.fa aln.bam > aln.cram o Convert a BAM file to a CRAM with NM and MD tags stored verbatim rather than calculating on the fly during CRAM decode, so that mixed data sets with MD/NM only on some records, or NM calculated using different definitions of. In a BAM file, each column of information is stored in its native data type (i.e. o Convert a BAM file to a CRAM file using a local reference sequence. the entirety of the query is stored in the aligned BAM, but the CIGAR field. This is called a BAM (Binary Alignment Map) file. USEARCH can read CIGAR strings using this operation, but does not generate them.Īlignment column containing a mismatch, i.e. PacBio-produced BAM files are fully compatible with the BAM specification. In this case, H operations specify segments at the start and/or end of the query that do not appear in the SAM record.Īlignment column containing two identical letters. This is used with hard clipping, where only the aligned segment of the query sequences is given (field 10 in the SAM record).
modern sam specification and by default will output cigar strings with X and. Segment of the query sequence that does not appear in the alignment. Output formats are fasta, fastq, sam, or bam (if samtools is installed). Filter a VCF file annotated with SNPEff or VEP with terms from Sequence-Ontology. Filters a BAM using a javascript expression ( java nashorn engine ). In this case, S operations specify segments at the start and/or end of the query that do not appear in a local alignment. Jvarkit : Java utilities for Bioinformatics. This is used with soft clipping, where the full-length query sequence is given (field 10 in the SAM record). Segment of the query sequence that does not appear in the alignment. USEARCH generates CIGAR strings containing Ms rather than X's and ='s (see below). This could contain two different letters (mismatch) or two identical letters.

Match (alignment column containing two letters).


 0 kommentar(er)
0 kommentar(er)
